1 –Technical note from Reichert Technologies, 3362 Walden Ave., Suite 100, Depew, NY 14043
INTRODUCTION
Surface Plasmon Resonance (SPR) is an invaluable technique that generates information-rich data for a variety of biomolecular interactions. Researchers utilize SPR to understand biological pathways and to develop and characterize a range of potential therapeutics to treat disease and other illnesses. In the context of the current coronavirus global pandemic, SPR can be employed to develop potential therapeutics to treat COVID-19 disease and help engineer vaccines to prevent virus infections. It can reveal vital structural and function information between the pathogen and host immune system that is critical in therapeutic development. SPR can be used to explore a variety of potential therapeutics that include low molecular weight compounds or drugs, peptides, proteins, and antibodies.
REICHERT SPR IN MALARIA VACCINE DEVLOPMENT
The importance of using SPR in guiding vaccine development is highlighted in two recent articles where researchers used a Reichert SPR system to investigate Malaria treatment.1,2 An integral step in malaria vaccine development has been to understand how infection by parasites occurs. Of the known human malaria parasites, the group whose research is highlighted here has targeted two – Plasmodium falciparum (P. falciparum)1, which is the most deadly parasite, and Plasmodium vivax (P. vivax)2 which is the most common parasite species that infects humans. A key question that needed to be answered early on is – How do parasites gain entry into red blood cells? This group discovered that P. vivax enters young red blood cells by making proteins that recognize and bind to receptors on the young red blood cell surface. This family of proteins was structurally similar to those used by P. falciparum to infect red blood cells. The next step in their research effort was to determine the three-dimensional structure of the proteins. This determination showed that the proteins are folded in the same way with the main difference being in the electrical charge on the surface of the molecules.
Researchers found that P. falciparum reticulocyte binding protein-like homologue 4 (PfRh4) binds to complement receptor 1 (CR1 or CD35) to mediate entry of malaria parasites into human red blood cells1. CR1 (complement receptor 1) is composed of 28–30 structural modules (which are called complement control protein (CCP) domains) in the extracellular domain. The CR1 protein fragments in the study outlined here consisted of only two CCP domains and had molecular weights of around 14,000 Daltons. Initial mapping studies identified the first two to three N-terminal modules of CR1 (CCPs 1-2 or CCPs 1–3) as specific inhibitors of the PfRh4-CR1 invasion pathway1. Seven different mutant proteins were tested for their inhibitory potential via SPR. SPR was able to rank them in terms of affinity with three having the highest affinity for the intended target. The SPR results matched with the expected biological activity determined independently with other techniques1. Binding affinities for a series of protein fragments to PfRh4 were determined. An example of responses from one of the higher affinity fragments is shown in Figure 1 (CR1 CCP 1-2). Data is fit to a 1:1 binding model with mass transport model:
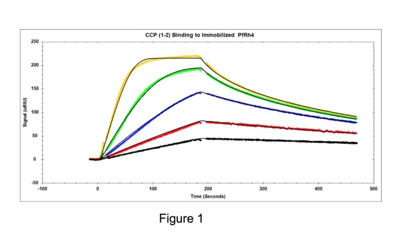
Figure 1: The following concentrations of the CR1 CCP1-2 inhibitor were injected over immobilized PfRh4: 5, 10, 20, 40 and 80 nM. Tracedrawer (Ridgeview Instruments AG) was used to fit the data. The calculated ka is 2.10 e6 M-1 s-1, the kd is 1.26 e-2 s-1, and the equilibrium dissociation constant KD is 6.01 nM.
This information is an important starting point that will lead to further screening of potentially more powerful inhibitors and can aid in the rational development of a novel malaria vaccine.
REICHERT SPR IN HIV/AIDS THERAPEUTIC DESIGN
Treatment with antiretroviral drugs is the accepted approach to HIV/AIDS therapy. Drugs are usually taken in combinations that include nucleoside reverse transcriptase inhibitors, which block how the HIV genetic material is used to create DNA from RNA, protease inhibitors, where the raw protein material for newly produced HIV viral particles is cut into specific pieces, and integrase inhibitors, which prevent the proviral HIV DNA from integrating into the host cell genome. These drugs do not provide a cure but significantly help slow the disease’s progression although with significant side-effects. Researchers in this current study (3) ultimately would like to develop a new class of antiretrovirals that can interfere specifically with the functions of HIV1/Nef. Since Nef is critical for HIV-1 replication in vivo and the immune escape of HIV-infected cells which leads to AIDS progression, development of potent HIV-1/Nef inhibitors hold promise in the suppression of HIV replication and the restoration of immune recognition of HIV-positive cells so that the patient’s own CTL response can be used to combat the disease.3 In this current work, Hydrogen Exchange Mass Spectrometry (HX MS) and SPR have been the primary techniques used to better understand the mechanism of action on Nef conformational transitions in solution by focusing on binding of Src-family kinases. Hence, this study supports the broader possibility that compounds interfering with conformational transitions in HIV-1/Nef structure have potential as a new class of antiretroviral agents.3 This is important as HIV can become resistant to existing treatments over time and new treatments will continue to be needed.
Researchers were interested in knowing whether binding properties changed depending on whether the Nef protein was full-length or truncated so that only the folded core lacking the N-terminal arm responsible for membrane anchoring (amino acids 1-60) was present. They also investigated a mutant defective for homodimer formation, in which Nef Asp123 was changed to Asparagine (Nef-D123N). Substitution of this highly conserved Aspartate prevented dimerization in cell-based fluorescence complementation assays and blocked important Nef functions related to infectivity and replication, as well as CD4 and MHC-I downregulation.3
After immobilization of SH3 or SH3-SH2 domains of Src-family kinases ligand, Nef protein was flowed over the chip either as full- length analyte, wild type core or D123N core. Examples of responses seen for the wild type Nef core are shown below in Figures 2 and 3. When SH3 alone was coupled, the wild type Nef core exhibited single-site binding with a KD value of of 128 nM. Binding was similar for the other Nef proteins – the affinity was a little lower for the full-length Nef (180 nM) and a little higher for the D123N core (119 nM). When SH3-SH2 was employed as the ligand, 2-site binding (conformational change) was seen with an affinity of 475 nM for the Nef core and similar affinities for the other Nef proteins (429 nM and 475 nM for the full-length Nef and D123N core, respectively). Hence, it was found that the mutation had little impact on the kinetics or affinity of SH3 domain interaction, suggesting that the D123N mutation does not influence the conformation of Nef to inhibit binding. This finding was consistent with HX MS results.3
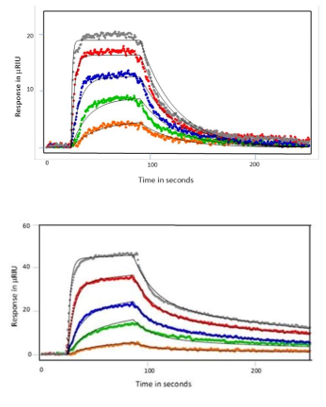 Figure 2: SH3 protein binding to Nef wild type core fit to a 1:1 binding model. Kinetic Rate constants determined in Tracedrawer are as follows: ka = 2.24 x 105 M-1s-1 and kd = 2.76 x 10-2 s-1 with an equilibrium dissociation constant (KD) of 128 nM.
Figure 2: SH3 protein binding to Nef wild type core fit to a 1:1 binding model. Kinetic Rate constants determined in Tracedrawer are as follows: ka = 2.24 x 105 M-1s-1 and kd = 2.76 x 10-2 s-1 with an equilibrium dissociation constant (KD) of 128 nM.
Figure 3: SH3-SH2 proteins binding to Nef wild type core fit to a 2-state conformational change model. Kinetic rate constants determined in Tracedrawer are as follows: ka1 = 6.29×104 M-1s-1, kd1 =5.65×10-2 s-1 and ka2 =1.06×102 M-1s-1, kd2 = 4.72×10-3 s-1 with an equilibrium dissociation constant of KD of 475 nM.
REICHERT SPR IN MONOCLONAL THERAPEUTIC ANTIBODY DEVELOPMENT
Transmembrane 4 superfamily member 5 protein (TM4SF5) is an important possible target for hepatocellular carcinoma and colon cancer. In this work, researchers utilized a cyclic peptide that mimics a structural motif of TM4SF5 as an antigen and then developed a monoclonal antibody that recognizes the TM4SF5 protein.4 The researcher employed Reichert SPR to characterize the antibody and showed that it has a very slow off rate making it an attractive antibody for potential therapeutic outcome. In this study, it was shown that both the cyclic vaccine and the humanized anti-TM4SF5 antibody suppressed the formation and growth of lung metastases that were established through intravenous injection of colon cancer cells in a mouse metastases model.4
CONCLUSION
SPR can play a pivotal role in therapeutics development and vaccine design. Reichert’s SPR systems have been implemented in a variety of research application (see https://www.reichertspr.com/publications/) and have the sensitivity and performance characteristics to meet challenging experimental needs. Reichert offers two main SPR platforms, the 2SPR and 4SPR systems that are affordable, flexible and have outstanding performance. Please contact us to learn more about how Reichert SPR can help you with your therapeutic and vaccine research during this critical time.
REFERENCES
- Nicholas T. Y. Lim, Markus J. Harder, Alexander T. Kennedy, Clara S. Lin, Christopher Weir, Alan F. Cowman, Melissa J. Call, Christoph Q. Schmidt and Wai-Hong Tham, Characterization of Inhibitors and Monoclonal Antibodies That Modulate the Interaction between Plasmodium falciparum Adhesin PfRh4 with Its Erythrocyte Receptor Complement Receptor 1,” J. Biol. Chemistry (2015) Vol. 290, pp. 25307-25321, doi:10.1074/jbc.M115.657171.
- Jakub Gruszczyk, Usheer Kanjee, Li-Jin Chan, Sébastien Menant, Benoit Malleret, Nicholas T. Y. Lim, Christoph Q. Schmidt, Yee-Foong Mok, Kai-Min Lin, Richard D. Pearson,,10 Gabriel Rangel, Brian J. Smith, Melissa J. Call, Michael P. Weekes, Michael D. W. Griffin, James M. Murphy, Jonathan Abraham, Kanlaya Sriprawat, Maria J. Menezes, Marcelo U. Ferreira, Bruce Russell, Laurent Renia, Manoj T. Duraisingh, Wai-Hong Tham1, “Transferrin receptor 1 is a reticulocyte-specific receptor for Plasmodium vivax,” Science (2018), Volume 359, pp. 48-55.
- Jamie A. Moroco, John Jeff Alvarado, Ryan P. Staudt, Haibin Shi, Thomas E. Wales, Thomas E. Smithgall, John R. Engen, “Remodeling of HIV-1 Nef Structure by Src- Family Kinase Binding,” Journal of Molecular Biology, Available online 16 December 2017, ISSN 0022-2836, doi.org/10.1016/j.jmb.2017.12.008.
- Guang Wu, Dongbum Kim, Byoung Kwon Park, Sangkyu Park, Ji-Hee Ha, Te Ha Kim, Avishekh Gautam, Jung Nam Kim, Su In Lee, Han-Bum Park, Yong-Sung Kim, Hyung-Joo Kwon, Younghee Lee, “Anti-metastatic effect of the TM4SF5-specific peptide vaccine and humanized monoclonal antibody on colon cancer in a mouse lung metastasis model,” Oncotarget (2016), Volume 7, pp. 79170 – 79186.


